Zaborsky Group
Clonal heterogeneity and immune changes in cancer

Research topic
TEAM Zaborsky uses various single-cell techniques to study cancer cells and their interactions with immune cells. In addition to tumor heterogeneity and the clonal evolution of cancer cells, the therapy response in patients and in the mouse model is also analyzed, as well as its connection with the intestinal microbiome.
Our focus
- Clonal evolution and tumor heterogeneity (single-cell genomics)
- Anti-cancer immunity and interactions between immune cells
- Immunophenotyping
- Interaction between gut microbiota and immune cells
- Therapy response and resistance mechanisms

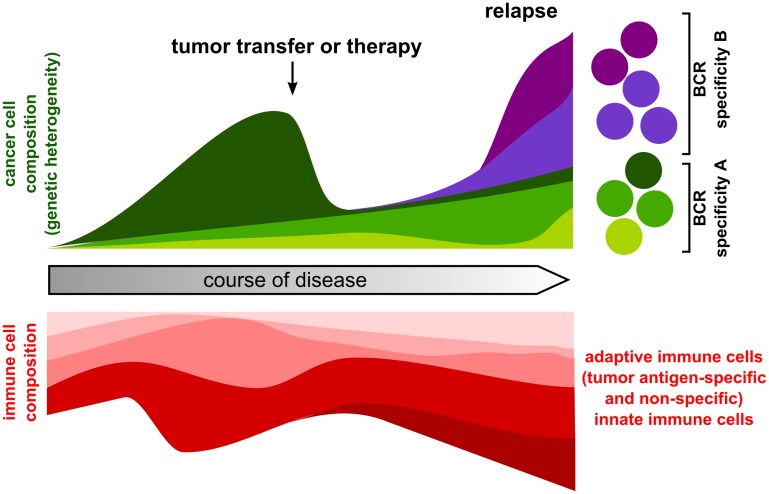
Malignant cells are in constant interaction with immune cells, creating a two-way selection pressure that shapes subclonal cancer evolution and leads to a specific change in the composition of immune cells that determines the success or failure of immune surveillance and therapies.
Tumor heterogeneity and clonal evolution
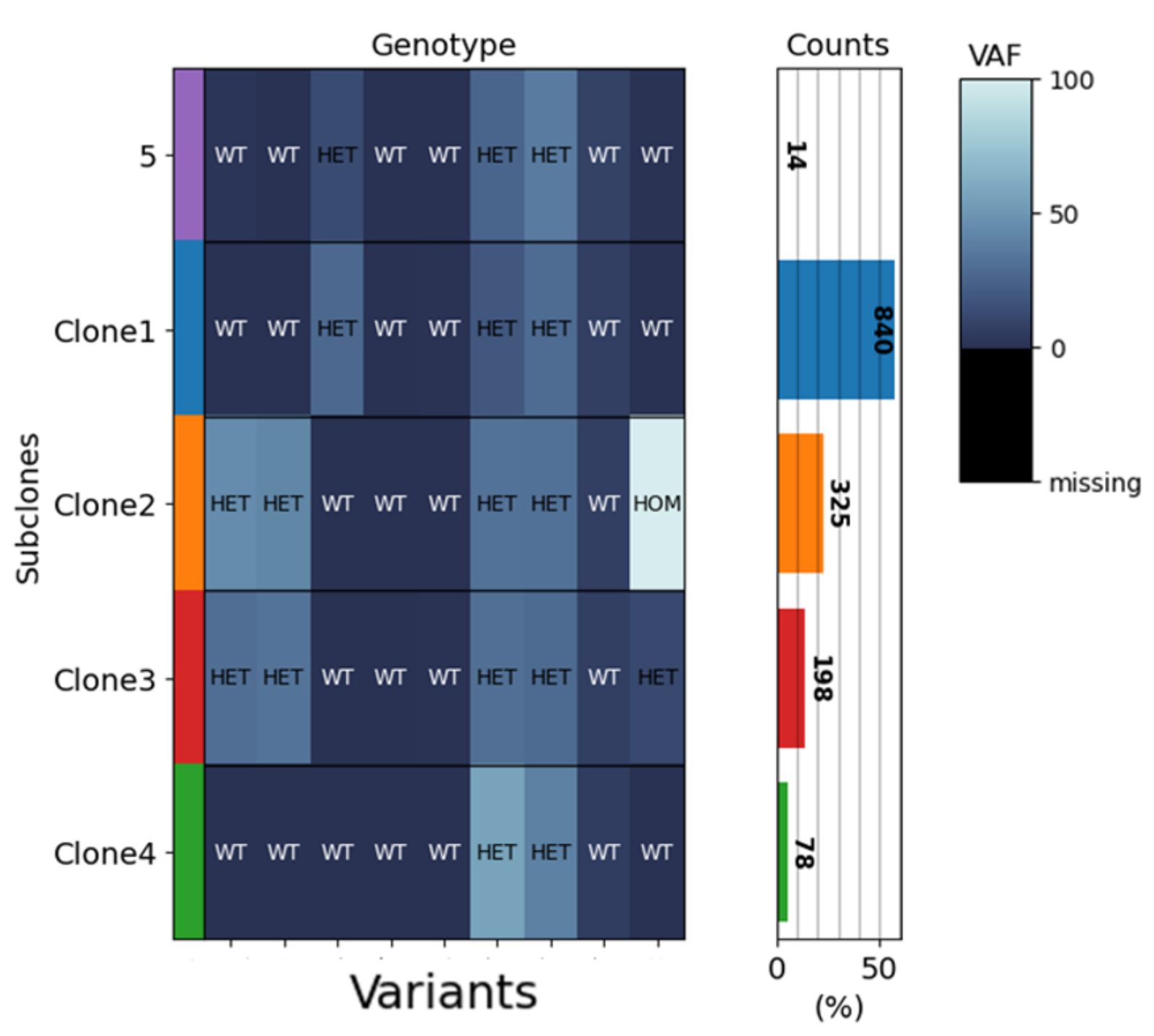
Chronic lymphocytic leukemia (CLL), a very common B-cell leukemia in older people, is characterized by the accumulation of non-functional B-cells in lymph nodes and peripheral blood. High-throughput sequencing approaches on several cancer entities have revealed complex genetic landscapes in human cancers. A complex set of clonal and subclonal mutations has also been described for CLL. The heterogeneity of tumors can play an important role in the diagnosis and treatment of cancer and can be decisive for the response to treatment.
Better understanding of cancer-immune interactions
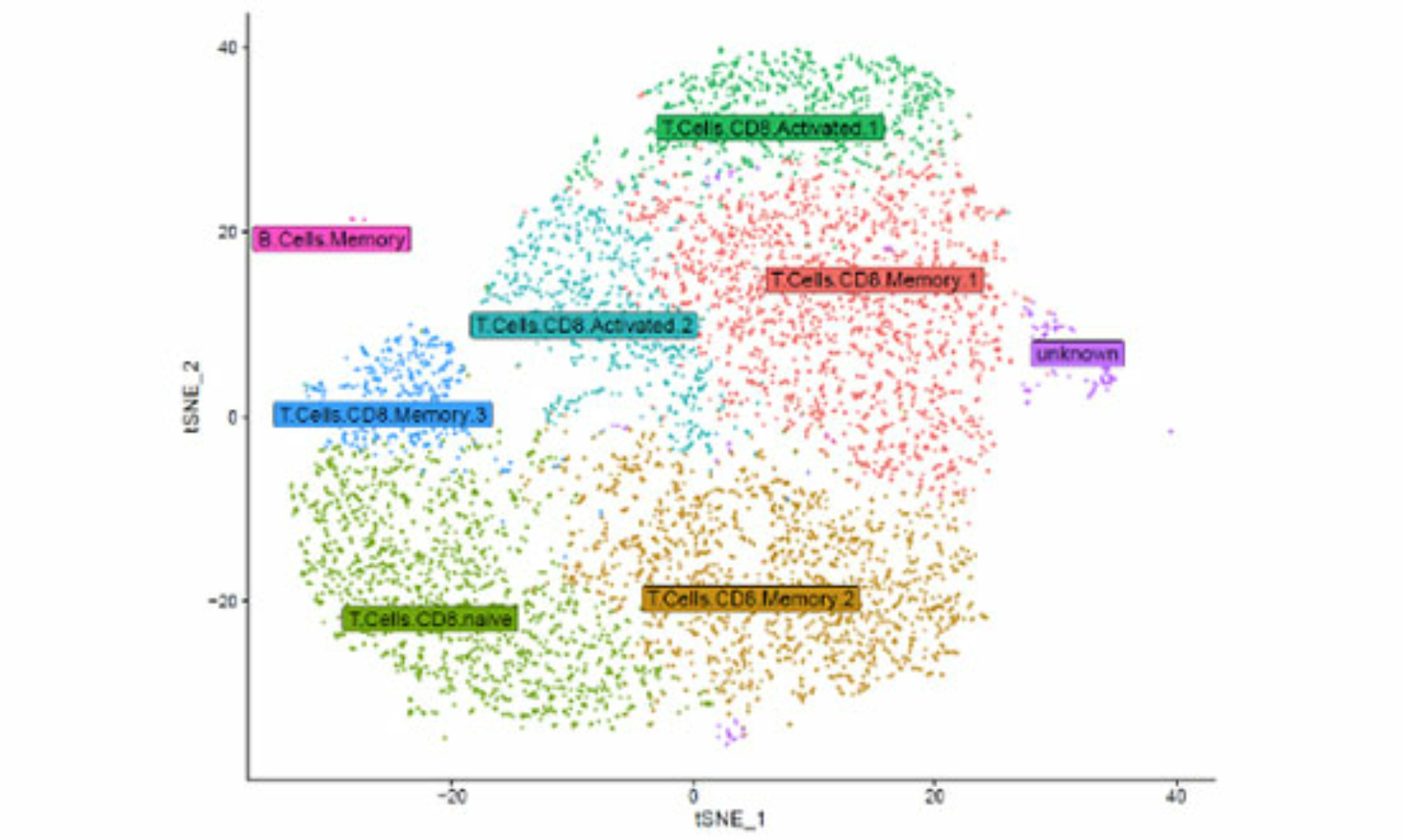
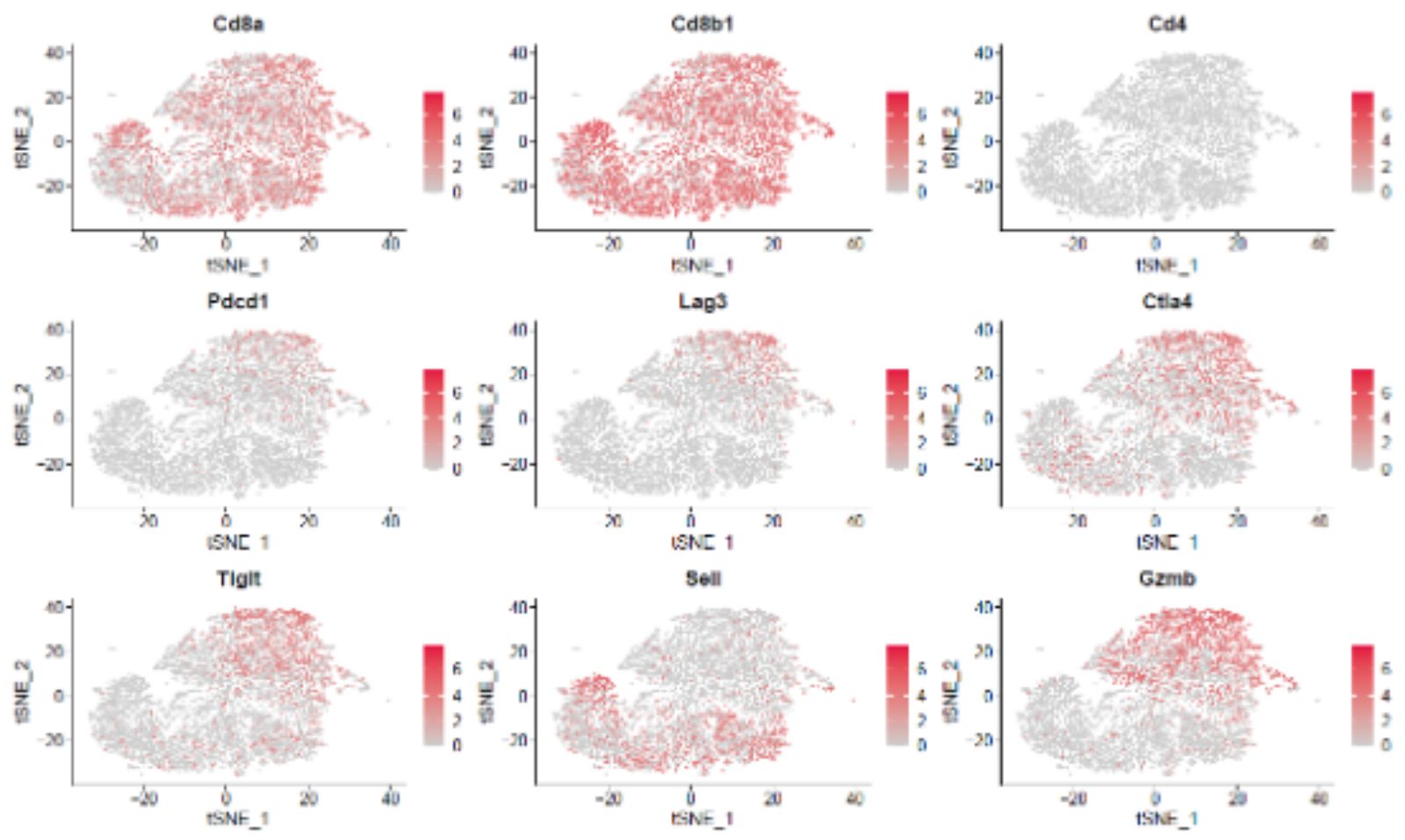
We use different mouse models to gain insights into the complex tumor biology. Among other things, we use the CLL mouse model (TCL1tg) to investigate the reorganization of the microenvironment and the response to immunotherapies. We are also interested in CLL-immune interactions in different organs. Through the phenotypic and functional characterization of (potentially tumor antigen-specific) T cells, we aim to better understand the anti-cancer immune response in CLL and attempt to reactivate immune cells.
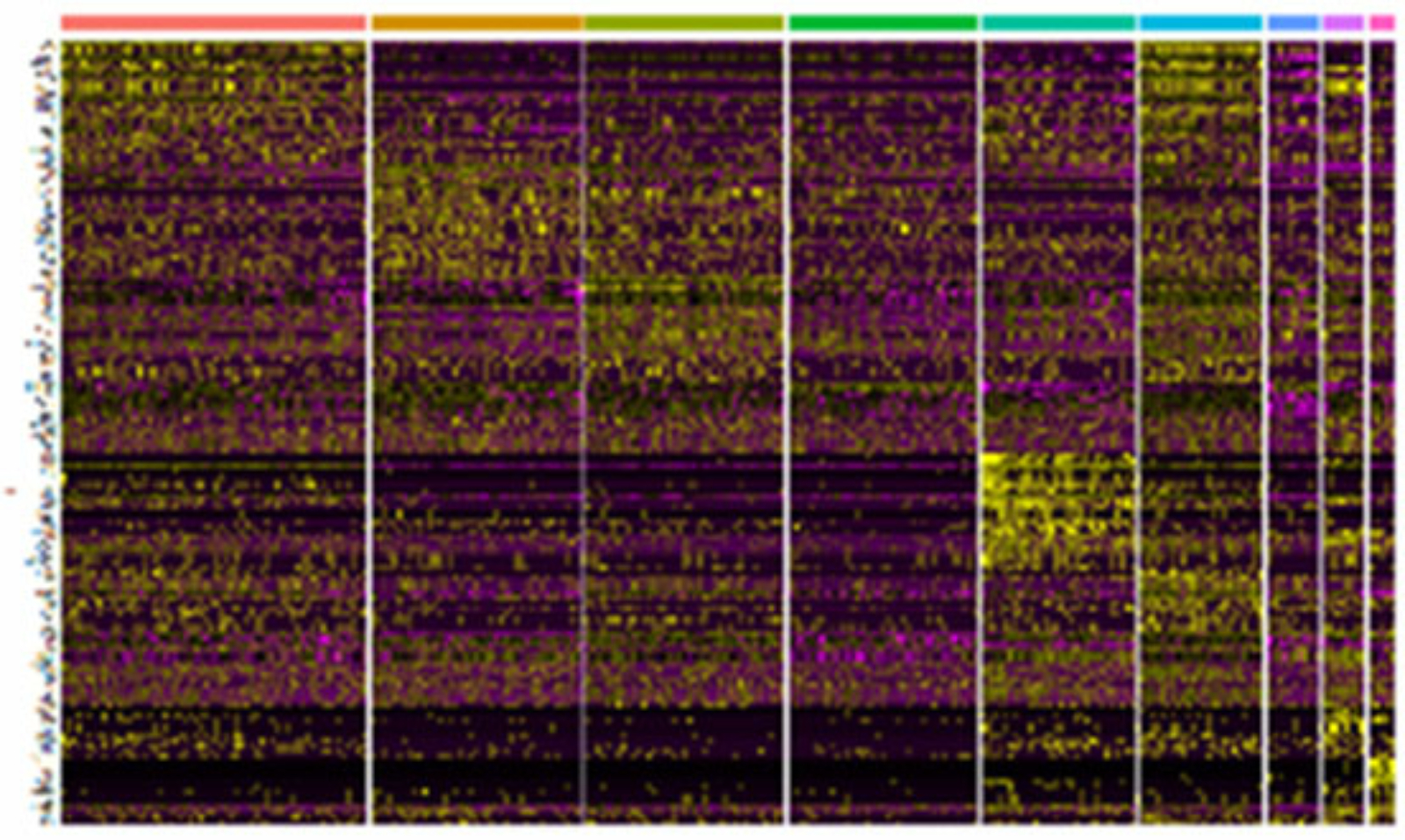
“The study of cancer-immune interactions is of paramount importance, not only to understand cancer progression and immune evasion, but also to improve current immune checkpoint therapy approaches, which – although considered a breakthrough in cancer therapy – are not effective in CLL and some other cancer entities.”
Priv.-Doz. Dr. Nadja Zaborsky
Intestinal microbiota and its interactions with immune cells
Since the gut microbiota is crucial for the response to immunotherapies, we are investigating the taxonomic composition in cancer patients and in various mouse models. We are also interested in the interaction of the microbiota with the various immune cells in the gut. A better understanding of this interaction could be used in the future to modulate the composition of the gut microbiota in order to improve the patient’s response to immunotherapies.
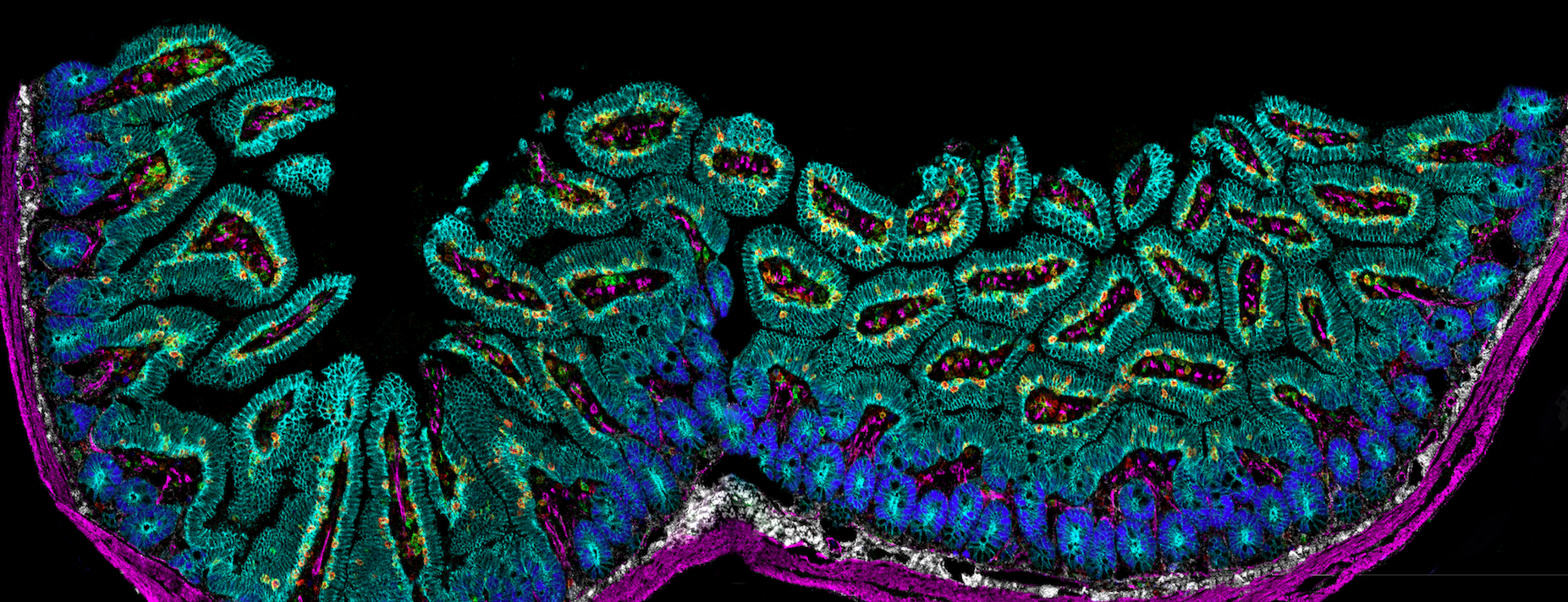

Imaging mass cytometry of immune cells in the small intestine.
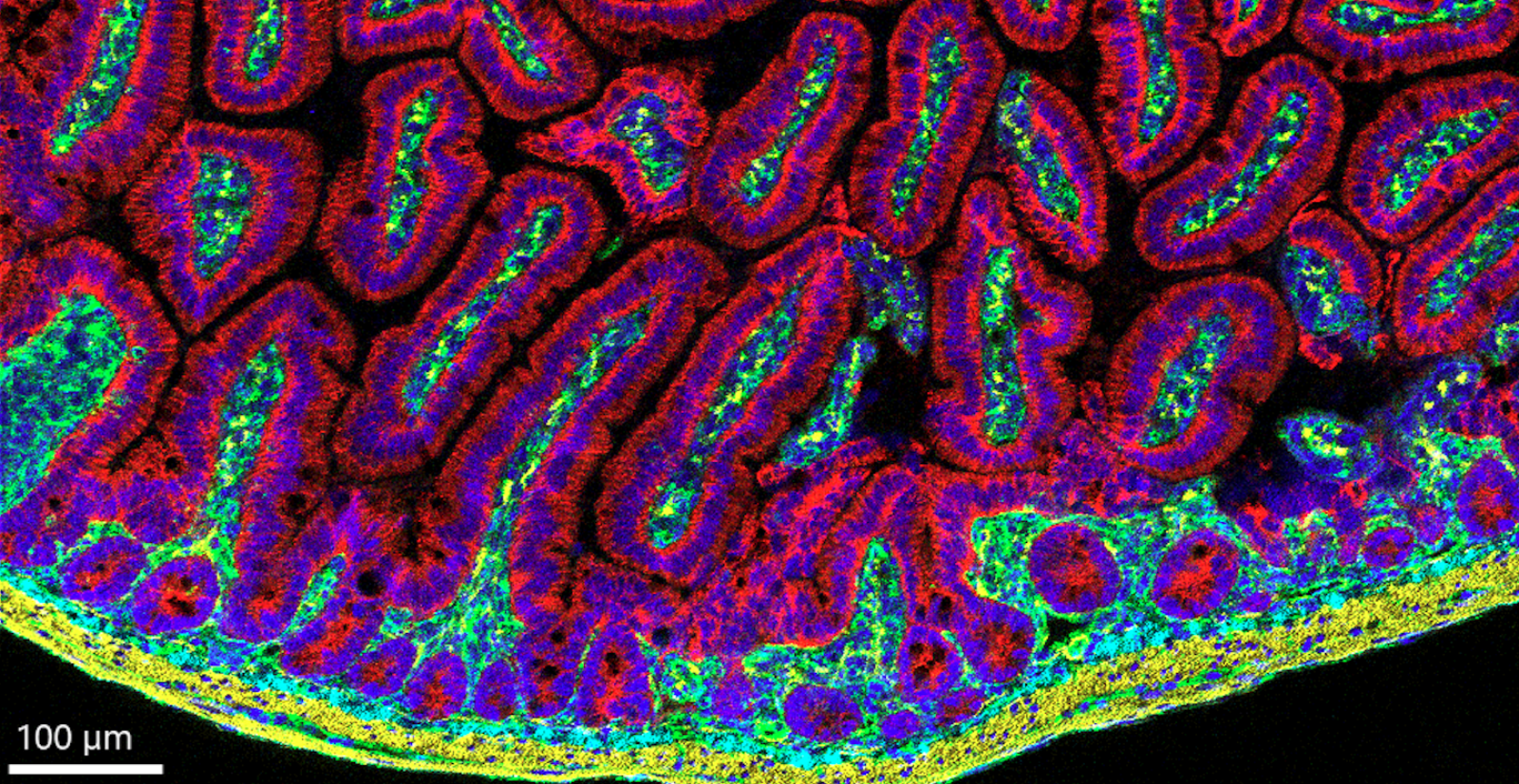
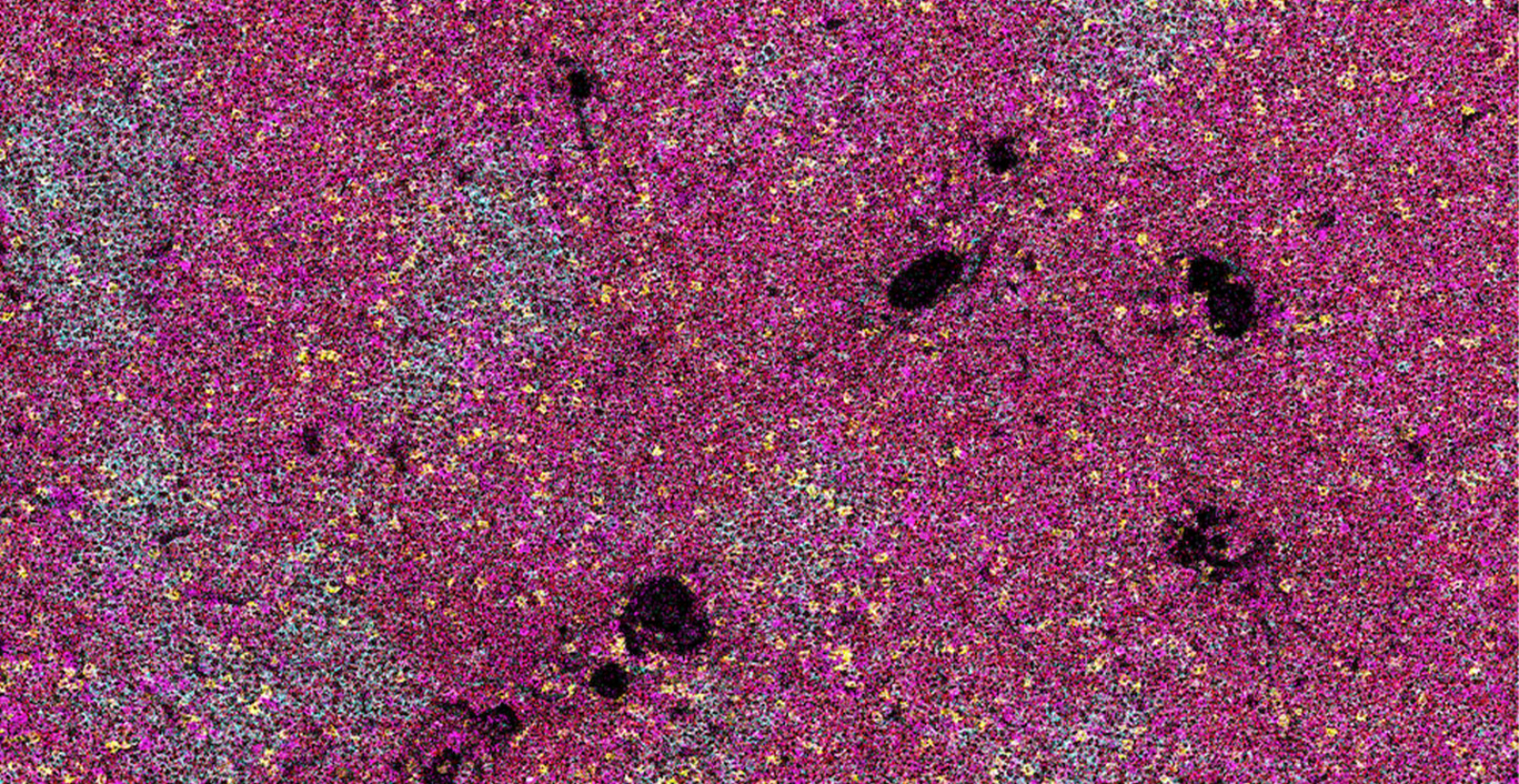
Imaging mass cytometry: leukemia-infiltrated lymph node (left) and CLL free after treatment (right).

In this lab, WE BELIEVE
https://sammykatta.com/diversity
(adapted with biorender)

Team
Nadja Zaborsky
(Group leader)
https://orcid.org/0000-0002-9775-185X
Thomas Parigger
(Post-Doc)
Aryunni Abu Bakar
(PhD student)
Stephan Drothler
(PhD student)
Christian Scherhäufl
(PhD student)
Jan Höpner
(PhD student)
Omar Mohamed
(Master student)
News:
CCS-CLL Workshop 2023


Publications
Parigger T, Gassner FJ, Scherhäufl C, Bakar AA, Höpner JP, Hödlmoser A, Steiner M, Catakovic K, Geisberger R, Greil R, Zaborsky N. Evidence for Non-Cancer-Specific T Cell Exhaustion in the Tcl1 Mouse Model for Chronic Lymphocytic Leukemia. Int J Mol Sci. 2021 Jun 22;22(13):6648. doi: 10.3390/ijms22136648.
Steiner M, Gassner FJ, Parigger T, Neureiter D, Egle A, Geisberger R, Greil R, Zaborsky N. A POLE Splice Site Deletion Detected in a Patient with Biclonal CLL and Prostate Cancer: A Case Report. Int J Mol Sci. 2021 Aug 30;22(17):9410. doi: 10.3390/ijms22179410.
Parigger T, Greil R, Zaborsky N. Mouse models to decipher anti-tumor immunity. Oncotarget. 2019 Aug 20;10(49):5005-5006. doi: 10.18632/oncotarget.27111. eCollection 2019 Aug 20.
Zaborsky N, Gassner FJ, Höpner JP, Schubert M, Hebenstreit D, Stark R, Asslaber D, Steiner M, Geisberger R, Greil R, Egle A. Exome sequencing of the TCL1 mouse model for CLL reveals genetic heterogeneity and dynamics during disease development.Leukemia. 2019 Apr;33(4):957-968. doi: 10.1038/s41375-018-0260-4.
Zaborsky N, Gassner FJ, Asslaber D, Reinthaler P, Denk U, Flenady S, Hofbauer JP, Danner B, Rebhandl S, Harrer A, Geisberger R, Greil R, Egle A. CD1d expression on chronic lymphocytic leukemia B cells affects disease progression and induces T cell skewing in CD8 positive and CD4CD8 double negative T cells. Oncotarget. 2016 Aug 2;7(31):49459-49469. doi:10.18632/oncotarget.10372.
Do you have a request or questions?
Get in touch with us – we will get back to you as soon as possible.