Group Egle
Immune interactions and signaling pathways in human and murine CLL

Concepts/basics
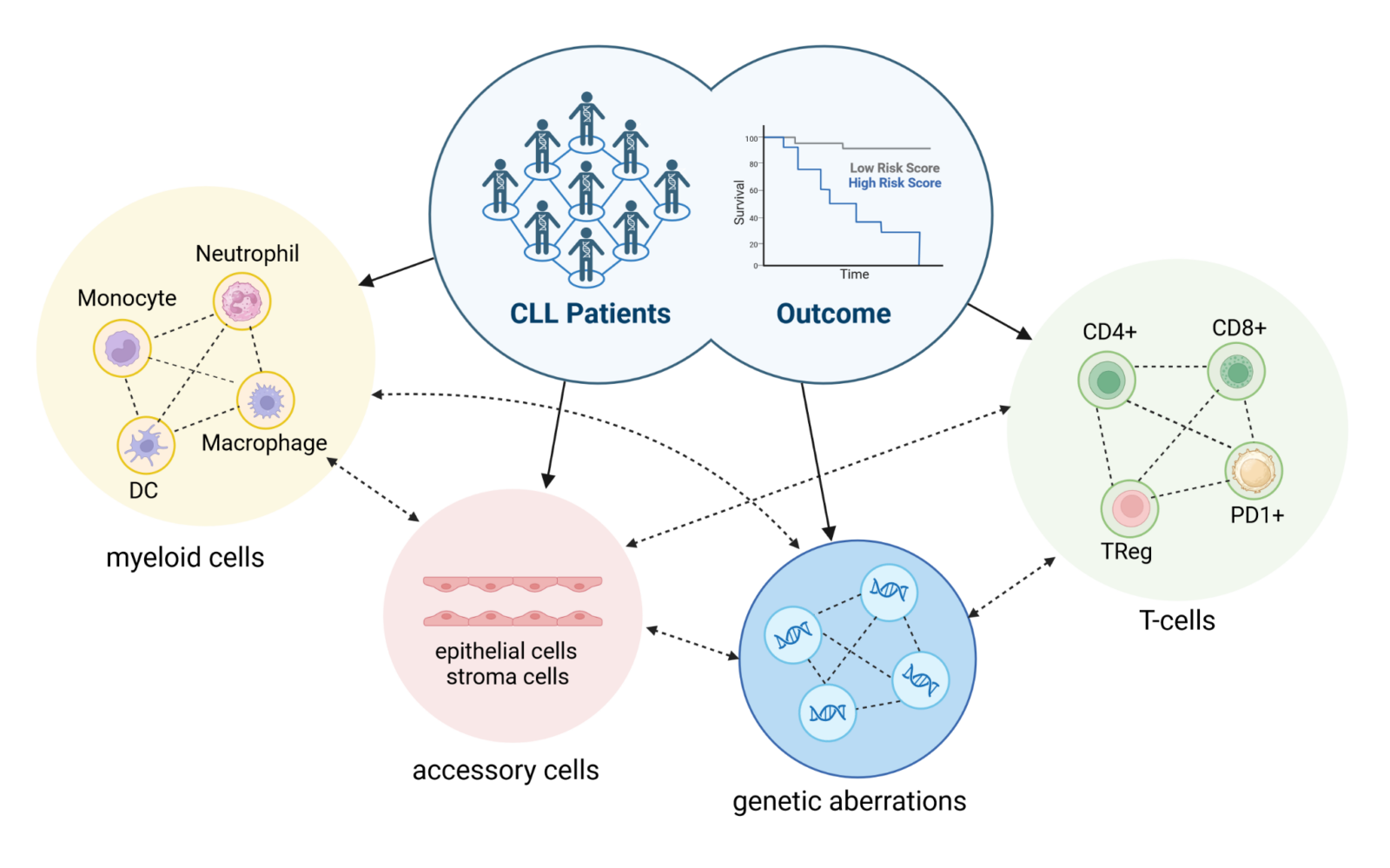
Tumor biology is a complex interplay of changes in cells that enable the clonal evolution of a self-replicating organism and do not adapt to the global goals of the organism. In addition to changes in the genetic profile due to mutation or epigenetic dysregulation, there are numerous cellular interactions (e.g. with the niche environment, the immune system, etc.) and programmed restrictions (e.g. cell stage-specific expression and signaling systems as well as identities), the evolution of which enables the development of a self-replicating tumor(Figure 1).
Chronic lymphocytic leukemia (CLL ) is not only an extremely relevant and stressful disease for our patients in the clinic, but also a unique model for studying the complex interactions described above. It has a complex and heterogeneous genetic landscape, defined interactions with the microenvironment and the immune system as well as specific links at the developmental level of the tumor cells. These different facets of the disease can be investigated in order to better understand the tumor biology and thus make it usable for therapeutic concepts.
Solution
The Egle group faces the complex task of modeling cellular signaling and cellular behavior in the context of numerous relevant cell-cell interactions, using an approach that leverages clinical access to validated primary clinical samples from patients in combination with state-of-the-art profiling technologies and complex culture approaches. In addition, the working group focuses on the development and implementation of
step-by-step validation of suitable mouse models for specific questions in order to enable clear proof-of-principle experiments. In this way, based on experiments with both primary patient samples and mouse models, we were able to explore and validate a number of important mechanisms that play a crucial role in the pathogenesis of CLL.
Progress/research results
Over the past 15+ years, the Egle group has developed mouse models for CLL using sophisticated genetic and experimental tools to gain new scientific insights in a variety of areas:
- Definition of essential signaling components involved in the development and maintenance of CLL, including initial proof-of-principle experiments in which the signaling components in the BCR signaling pathway were defined as valid therapeutic targets,1 a strategy that is currently undergoing clinical development of drugs for this signaling pathway.
- Definition of essential interactions between the microenvironment and tumor cells2, especially with regard to therapeutically relevant target structures.3
- Definition of immune interactions and immune evasion strategies (a mechanism by which tumor cells evade attacks by the immune system) that promote or limit the development of CLL.4,5
- Definition of pathway signatures that are associated with the clinical presentation of CLL independent of mutational changes in a pathway and that describe tumor biology using pathway patterns.6
- Definition of molecular changes in the biology of murine CLL by high-throughput sequencing.7
Project
Based on many years of experience and equipped with a growing arsenal of technical tools and genetic resources (both in human and murine CLL), we have now entered a conceptual “proof-of-principle phase” in which we can determine recurring and interacting signaling pathway patterns of CLL and describe them as so-called “signalotypes”.
Research focus
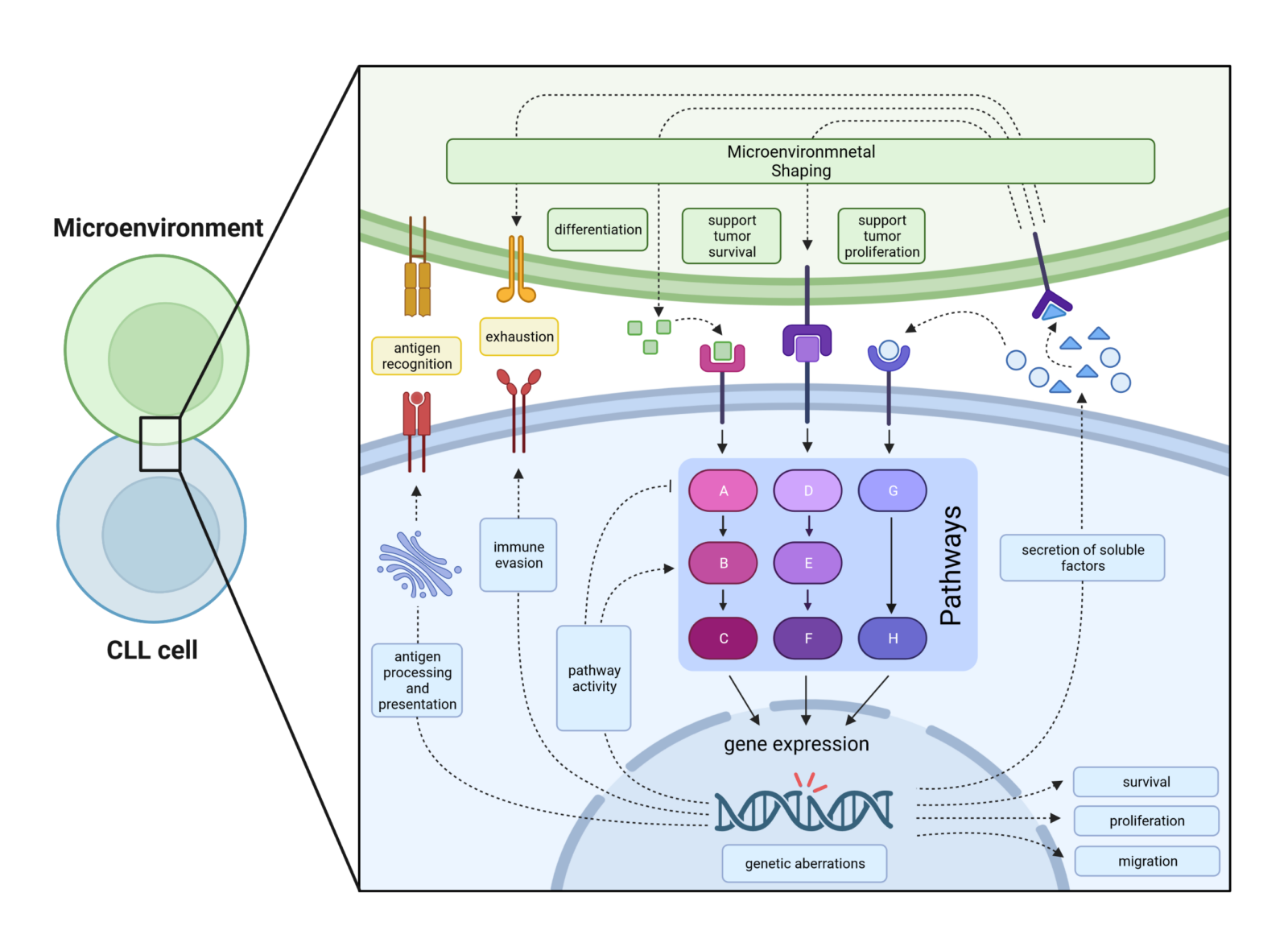
Chronic lymphocytic leukemia is a clinically and biologically extremely complex and heterogeneous disease that is characterized by chromosomal aberrations, mutations in various genes, the influence of the microenvironment2,8 and interactions with immune cells. The interaction of these factors ultimately has a strong impact on disease progression and treatment success(Figure 1).1,9,10 In CLL patients, disease heterogeneity is usually determined by classical prognostic factors, genetic aberrations and the percentage distribution of immune cell (sub)groups. In contrast, we pursue an innovative approach based on the hypothesis that the combination of all mentioned factors is reflected in CLL intrinsic pathway (de)regulation(Figure 2), which we analyze by combining CLL transcriptome and phosphoproteome data.
Furthermore, we model CLL heterogeneity in vivo by using different genetically modified mouse models on a Tcl-1 transgenic (tg) background(Figure 3).5 The Tcl-1 transgene is responsible for the development and establishment of CLL in the mouse model, while the genetic defects in known targets modulate the development of CLL or resistance to therapy by increasing the dependence on the microenvironment.3 and/or the communication of CLL cells with immune cells4 and ultimately alter the pattern of (de)regulation of the intrinsic CLL signaling pathway pattern.
In our studies, we use the Tcl-1 transgenic mouse model, which develops murine CLL that is very similar to human disease. Tcl1 transgenic animals are crossed with genetically modified mice that either have defects in apoptosis regulation (e.g. various members of the Bcl-2 family), in cell cycle regulation (e.g. p21), in signaling pathways (e.g. BIRC3, loss of ubiqutin ligase function) or in immune interactions (e.g. IRF4). In addition, mouse models with defects in cytokine signaling are planned. Using these different mouse models, we monitor CLL development over time, the architecture of B-cell receptors (which is considered one of the most important factors in CLL pathophysiology) and the composition or differentiation status of immune cells. This occurs both in primary models and as a result of tumor transplantation into immunocompetent or immunodeficient recipient mice(Figure 3).
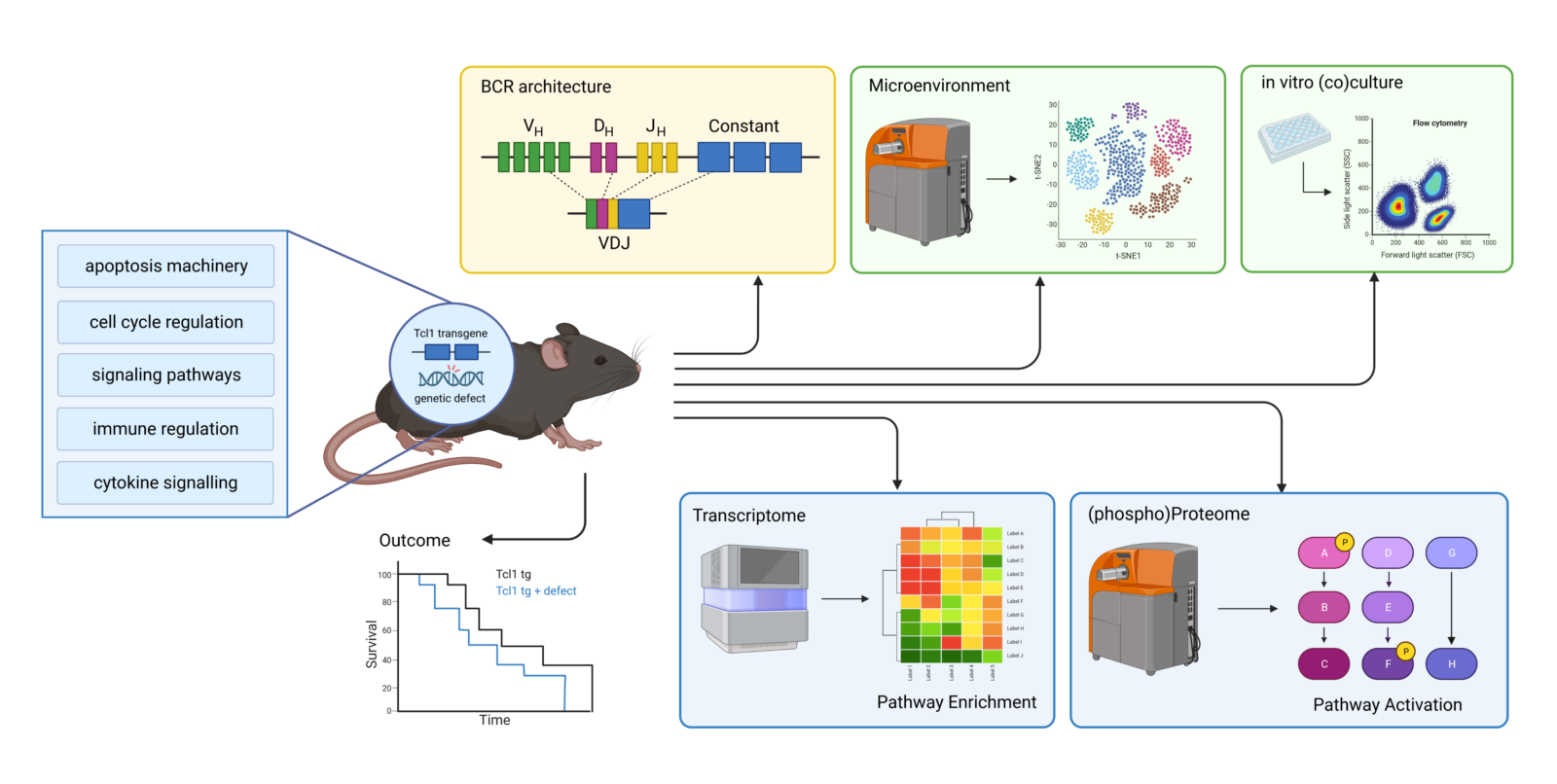
Monitoring CLL development and progression in vivo is of great importance as it is known that CLL tumor cells shape the surrounding microenvironment according to their needs. This protects the tumor cells from an intrinsic or recipient-induced anti-tumor immune response while providing survival-promoting and/or proliferation-enhancing factors that promote tumor growth. In addition, we determine the sensitivity of CLL cells after drug treatment or after stimulation of CLL cells in vitro. The transcriptome of CLL cells (with or without stimulation/treatment) is analyzed by RNA-Seq and the (phospho) proteome by single cell mass cytometry and classical methods (e.g. phospho-flow). Our aim is to combine data from these different sources: Superlensing time, B cell receptor architecture, transcriptome, (phospho) proteome and in vitro stimulation/treatment to bioinformatically combine and ultimately investigate signaling pathway patterns specific to a particular genetic defect. This includes the interaction of different signaling pathways and CLL’s own compensation for lost or gained signals, as well as the effects on the shaping of the microenvironment, the survival and growth potential of the tumor and the mechanisms for avoiding immune attacks.
Predicting the clinical course of CLL in terms of prognosis, treatment success and possible disease relapse is a challenge, as a number of different parameters are involved in the pathogenesis of CLL. This includes not only the genetic prerequisites (such as mutations or chromosomal aberrations), but also the architecture of the microenvironment and the interaction with other immune cells. As mentioned above, CLL cells shape their microenvironment to evade immune attacks by driving the differentiation of immune cells (e.g. into exhausted T cell subsets). In addition, CLL cells stimulate surrounding cells of the microenvironment to provide survival-promoting and/or proliferation-promoting factors that promote tumor growth. Furthermore, genetic defects can facilitate changes in the microenvironment and/or alter the expression or function of tumor-relevant genes(Figure 2). The wide variety of mutated genes and the fact that the same genes are only mutated in a very small percentage of CLL patients suggests that these mutations obviously favor the pathogenesis of CLL, but are not essential. However, if we look at the signaling pathways, we see that the high number of mutated genes is associated with a very limited number of affected signaling pathways. Therefore, we hypothesize that the CLL pathway pattern of individual patients, which represents a cumulative signal from the aberrant expression of tumor-relevant genes, either caused by genetic defects or interactions with the microenvironment, is a better parameter for clinical outcome than any single piece of information alone. Similar to the mouse model, we use a combination of information from the transcriptome, the (phospho)proteome and genetic aberrations to determine the pattern of the signaling pathways, which are combined bioinformatically and analyzed with the help of signaling pathway databases. Finally, the clinical results and the results of the in vitro stimulation/treatment are integrated.
Team
Alexander Egle (PI)
ORCID: 0000-0003-0648-4416
Daniela Asslaber (PostDoc)
ORCID: 0000-0002-8711-016X
Jennifer Luise Forster (PhD student)
ORCID: 0009-0000-5247-0512
Important publications
Asslaber D, Qi Y, Maeding N, et al. B-cell-specific IRF4 deletion accelerates chronic lymphocytic leukemia development by enhanced tumor immune evasion. Blood. 2019;134(20):1717-1729.
Asslaber D, Wacht N, Leisch M, et al. BIRC3 Expression Predicts CLL Progression and Defines Treatment Sensitivity via Enhanced NF-kappaB Nuclear Translocation. Clin Cancer Res. 2019;25(6):1901-1912
Zaborsky N, Gassner FJ, Hopner JP, et al. Exome sequencing of the TCL1 mouse model for CLL reveals genetic heterogeneity and dynamics during disease development. Leukemia. 2018.
Egle A, Steurer M, Melchardt T, et al. Fludarabine and rituximab with escalating doses of lenalidomide followed by lenalidomide/rituximab maintenance in previously untreated chronic lymphocytic leukaemia (CLL): the REVLIRIT CLL-5 AGMT phase I/II study. Ann Hematol. 2018;97(10):1825-1839.
Kocher T, Asslaber D, Zaborsky N, et al. CD4+ T cells, but not non-classical monocytes, are dispensable for the development of chronic lymphocytic leukemia in the TCL1-tg murine model. Leukemia. 2016;30(6):1409-1413.
Greil R, Obrtlikova P, Smolej L, et al. Rituximab maintenance versus observation alone in patients with chronic lymphocytic leukaemia who respond to first-line or second-line rituximab-containing chemoimmunotherapy: final results of the AGMT CLL-8a Mabtenance randomized trial. Lancet Haematol. 2016;3(7):e317-329.
Lutzny G, Kocher T, Schmidt-Supprian M, et al. Protein kinase c-beta-dependent activation of NF-kappaB in stromal cells is indispensable for the survival of chronic lymphocytic leukemia B cells in vivo. Cancer Cell. 2013;23(1):77-92.
Asslaber D, Grossinger EM, Girbl T, et al. Mimicking the microenvironment in chronic lymphocytic leukaemia – where does the journey go? BrJHaematol. 2013;160(5):711-714.
Hofbauer JP, Heyder C, Denk U, et al. Development of CLL in the TCL1 transgenic mouse model is associated with severe skewing of the T-cell compartment homologous to human CLL. Leukemia. 2011;25(9):1452-1458.
Do you have a request or questions?
Get in touch with us – we will get back to you as soon as possible.